NCI, Pawsey, and ARDC help researchers rapidly identify therapeutic phages from metagenomic data through the powerful new Sphae toolkit.
Antibiotic resistance is one of the most pressing global health threats of our time, prompting scientists to urgently explore alternative ways to combat deadly bacterial infections. One such promising solution lies in phage therapy using bacteriophages (viruses that infect and kill bacteria) to target specific pathogens. However, finding the right phage to treat a bacterial infection is like searching for a needle in a microbial haystack. Thanks to a powerful new computational toolkit called Sphae, developed by Bhavya Papudeshi and Prof. Robert Edwards from Flinders University (Australia), along with other researchers from The University of Adelaide (Australia), University of San Diego (USA) and the University of California San Diego (USA), that search just got faster, smarter, and more scalable.
Supported by the National Computational Infrastructure (NCI)’s Gadi supercomputer, Pawsey’s Setonix supercomputer, and Australian Research Data Commons (ARDC) Nectar Infrastructure — all enabled through the Australian Government's National Collaborative Research Infrastructure Strategy (NCRIS) program — the researchers have designed an automated pipeline that sifts through complex metagenomic sequencing data to uncover the best phage candidates for therapeutic use. The work has been published in the journal Bioinformatics Advances and represents a major step forward in personalised, data-driven phage therapy.
What Is Phage Therapy and Why Does It Matter?
Phage therapy harnesses naturally occurring viruses that infect and destroy specific strains of bacteria without harming human cells. Unlike broad-spectrum antibiotics, which can disrupt the body’s microbiome and lead to resistance, phages offer highly targeted precision treatment. This Phage therapy is particularly useful in treating drug-resistant infections where conventional antibiotics may no longer work.
However, designing an effective phage treatment requires identifying which phages can infect a particular bacterial strain—an incredibly complex process when working with metagenomic samples that contain thousands of unknown viral sequences.
Introducing Sphae: Automating the Discovery of Therapeutic Phages
Sphae is an end-to-end, modular toolkit designed to automate the prediction of phage therapy candidates from complex sequencing data. It combines genome assembly, taxonomic classification, quality checks, infection potential scoring, and host range prediction—all in one integrated workflow.
The pipeline filters and scores potential phages based on quality and clinical relevance, allowing researchers to quickly triage which viruses are suitable for therapeutic development. Importantly, Sphae doesn’t require manual annotations or reference genomes, which makes it adaptable to real-world clinical and environmental samples with unknown or highly variable genetic content.
How Gadi Supercharged the Sphae Toolkit
Running a pipeline as comprehensive as Sphae requires significant computational power, particularly when processing metagenomic datasets that can span hundreds of gigabytes. The team leveraged NCI’s Gadi supercomputer to perform large-scale testing and optimisation of the Sphae pipeline across a variety of synthetic and real-world samples.
Gadi enabled parallelisation and speed-up of multiple steps within the workflow—especially genome assembly, quality scoring, and host-range prediction—reducing analysis times from days to hours. This high-throughput capacity was key in validating Sphae across simulated infections, clinical samples, and publicly available phage therapy datasets.
The researchers used Gadi’s compute-intensive nodes to benchmark Sphae’s performance, demonstrating that it could recover the correct therapeutic phage in all test cases with high specificity and minimal false positives.
NCI’s Role in Empowering Bioinformatics Innovation
Australia’s leading high-performance computing (HPC) systems, including NCI’s Gadi supercomputer, the Pawsey Supercomputing Research Centre, and the Nectar Research Cloud, enabledthe development and testing of Sphae. For each major release, the Sphae workflow was rigorously validated across these platforms to ensure seamless performance in diverse computing environments. This comprehensive testing strategy was crucial for refining Sphae’s usability and reproducibility, making it more accessible to researchers working on phage genome assembly and annotation.
Recognising that HPC systems often differ in configurations—such as hardware architectures, software environments, and security protocols— NCI’s Gadi was utilised as a testing environment. As one of Australia’s most powerful supercomputing facility, Gadi is widely utilised by over 5,000 researchers across the country. By validating Sphae on Gadi, the researchersensured its robustness and compatibility within the diverse HPC environments commonly used by the Australian research community.
The Bigger Picture: A New Frontier in Precision Antimicrobial Treatment
Sphae enables rapid and scalable prediction of therapeutic phages from metagenomic data, opening doors to more personalised and accessible antimicrobial treatments. Its modular design allows it to evolve with advances in phage biology and adapt to diverse data types or clinical needs.
As antibiotic resistance grows, tools like Sphae will be critical for timely phage therapy interventions—potentially saving lives in both clinical and field settings.
This work highlights the essential role of supercomputing in biomedical innovation. Without Gadi’s power and scalability, developing and validating Sphae at this pace would not have been possible.
By supporting tools like Sphae, NCI continues to empower research tackling global challenges—from public health to climate change—making the future of personalised infectious disease treatment a tangible reality.
You can read the full research paper here: Sphae: an automated toolkit for predicting phage therapy candidates from sequencing data
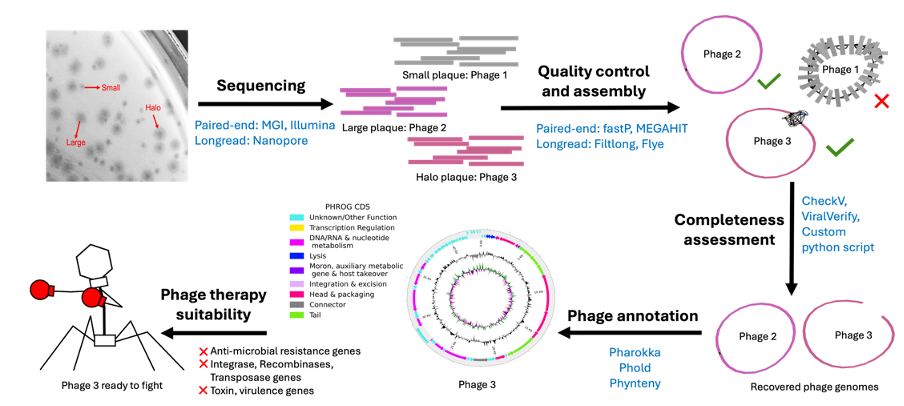
Figure: Flowchart of Sphae’s modular process for predicting suitable phages for antimicrobial therapy